The quest to fetch the details of the ever-elusive $Embedded$ connection to the local tabular data model has always interested the Power BI community. There are numerous tips on how to obtain the details involving either shell or PowerShell scripts or even V..B..A…
I want to share an approach based on using R.Execute() in Power Query and has the advantage of not implying a dependency outside the Power BI environment. This is a re-iteration of a pattern that I’ve already mentioned in one of my previous posts, which explained how to use R to get STDOUT as a data source in Power Query.
In the R script I use, the function system2() executes system commands tasklist and netstat. Because I need to output the results of these commands into data frames, I also use the function read.fwf() to parse the fixed delimited output of STDOUT.
| ##Get msmdsrv process details | |
| msmdsrv_proc <- system2("tasklist",args = c('/fi','"imagename eq msmdsrv.exe"'), stdout=TRUE) | |
| msmdsrv_clean<-msmdsrv_proc[-c(1,3)] | |
| msmdsrv<-read.fwf(textConnection(msmdsrv_clean),widths=c(25,9,17,12,13),comment.char="") | |
| ##Get all TCP ports | |
| tcp <- system2("netstat", args = c('-anop','TCP'), stdout=TRUE) | |
| ports<-read.fwf(textConnection(tcp),skip=3,widths=c(9,23,23,16,10)) |
In the Power Query world, I transform the two data frames msmdsrv and ports via function formatTable, which trims the table contents and promotes the headers. Then I match/join the two tables on [PID] and parse the contents of column [Local Address] to get the desired port number. Here is the whole script :
| let | |
| RScript = R.Execute("msmdsrv_proc <- system2(""tasklist"",args = c('/fi','""imagename eq msmdsrv.exe""'), stdout=TRUE) #(lf)msmdsrv_clean<-msmdsrv_proc[-c(1,3)]#(lf)msmdsrv<-read.fwf(textConnection(msmdsrv_clean),widths=c(25,9,17,12,13),comment.char="""")#(lf)tcp <- system2(""netstat"", args = c('-anop','TCP'), stdout=TRUE)#(lf)ports<-read.fwf(textConnection(tcp),skip=3,widths=c(9,23,23,16,10))"), | |
| formatTable = (T) => Table.PromoteHeaders(Table.TransformColumns(T,{{"V1",Text.Trim},{"V2",Text.Trim},{"V3",Text.Trim},{"V4",Text.Trim},{"V5",Text.Trim}})), | |
| msmdsrv = formatTable(RScript{[Name="msmdsrv"]}[Value]), | |
| ports = formatTable(RScript{[Name="ports"]}[Value]), | |
| match = Table.SelectRows(ports, each [PID]=Table.ToRecords(msmdsrv){0}[PID]){0}[Local Address], | |
| out = List.Last(Text.Split(match,":")) | |
| in | |
| out |
If you prefer keeping all the filtering logic inside the R script, here is my take on how to do it:
| let | |
| Source = #table(null,{{1}}), | |
| RScript = R.Execute("output<-dataset #(lf)msmdsrv_proc <- system2(""tasklist"",args = c('/nh','/fi','""imagename eq msmdsrv.exe""'), stdout=TRUE) #(lf)msmdsrv_pid <- strsplit(msmdsrv_proc[2],""\\s+"")[[1]][2]#(lf)msmdsrv_regex <- paste(c(""ESTABLISHED\\s*"", msmdsrv_pid),collapse="""")#(lf)tcp <- system2(""netstat"", args = c('-ano'), stdout=TRUE) #(lf)msmdsrv_connections<-grep(msmdsrv_regex,tcp,value=TRUE)#(lf)msmdsrv_inbound<-strsplit(strsplit(msmdsrv_connections[1],""\\s+"")[[1]][3],"":"")[[1]] #(lf)output$port<-tail(msmdsrv_inbound,n=1)",[dataset=Source]), | |
| out = RScript{[Name="output"]}[Value] | |
| in | |
| out |
As a final note, I wanted to add that there is actually a way of connecting to the embedded tabular data model, which doesn’t require any dependencies. Try the command:
= AnalysisServices.Databases("*")
Unfortunately, it uses the same connection context as the live connection, so you’ll get an exception like:
Microsoft.Mashup.Evaluator.Interface.ErrorException: Connections to the server context that is currently active are not supported.
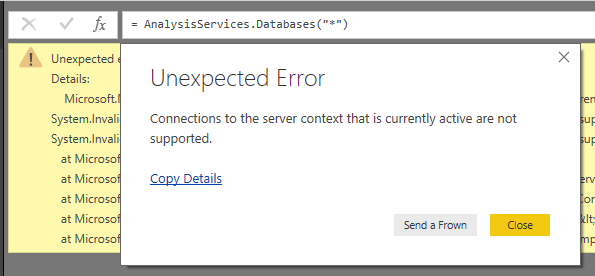
Which is not very practical.


2 Comments